The mechanism by which the conserved Ypt1/Rab1 GTPases mediate the beginning of two cellular processes, secretion and autophagy, is unknown. Gyurkovska et



Li Hao, Kuninori Suzuki et al. (University of Tokyo 東京大学大学院新領域創成科学研究科) report that lipophilic dye octadecyl rhodamine B (R18) is transported to the ER mediated by membrane transfer proteins. During autophagy, ER-resident R18 is transferred to the autophagic membrane via Atg2. After termination, R18 is reversed back to the ER, showing that the direction of bridge-type lipid transfer is modulated by metabolic states.
Bridge-like lipid transfer proteins (LTPs) contain a repeating β-groove domain and long hydrophobic grooves that act as bridges at membrane contact sites (MCSs) to efficiently transfer lipids. Atg2 is one such bridge-like LTP essential for autophagosome formation, during which a newly synthesized isolation membrane (IM) emerges and expands through lipid supply. However, studies on Atg2-mediated lipid transfer are limited to in vitro studies due to the lack of a suitable probe for monitoring phospholipid dynamics in vivo. Here, we characterized the lipophilic dye octadecyl rhodamine B (R18), which internalizes and labels the endoplasmic reticulum (ER) in a manner that requires flippases and oxysterol-binding protein–related proteins. Using R18, we demonstrated phospholipid transfer from the ER to the IM during autophagy in vivo. Upon autophagy termination, our data suggested the reversible phospholipid flow from the IM to the ER in response to environmental changes. Our findings highlight the critical role of bridge-like LTPs in MCS-mediated phospholipid homeostasis.

New research reveals that the Rhododendron pseudochrysanthum var. taitunense is a distinct subspecies, uniquely characterised by its glabrous leaves, larger seeds and specialised pollen morphology.
Found exclusively in the low-elevation mountains of Northern Taiwan, this rare taxon stands clearly apart from its close relatives within the species complex.
Read the full paper: doi.org/10.3897/phytokeys.271.
Our study examines the morphological and statistical differentiation within the Rhododendron pseudochrysanthum species complex through comparative analyses of macro-and micro-morphological characters. Using significance testing and cluster analysis, our results demonstrate that R. pseudochrysanthum Hayata ssp. morii (Hayata) Yamazaki var. taitunense Yamazaki is distinct from other members of the complex, namely R. morii Hayata, R. pseudochrysanthum Hayata, and R. hyperythrum Hayata. This taxon is characterized by glabrous mature leaves with revolute margins, larger flower buds with an elongated conical shape, larger pollen and seed sizes, and distinct pollen and seed morphology. Furthermore, R. pseudochrysanthum ssp. morii var. taitunense exhibits a restricted and localized distribution, occurring exclusively in low-elevation mountainous areas of Northern Taiwan.

Learn how the “rejuvenating” speed of light, antimatter propulsion, and wormholes could help speed up the vast distances of space during interstellar travel.
In this video, we explore one of the most fascinating concepts in interstellar travel: **generation ships**, massive spacecraft that would carry human civilizations across the stars, with journeys lasting hundreds or even thousands of years. As humanity dreams of traveling beyond the solar system, the idea of generation ships offers a possible solution to the immense distances between stars—a journey that could span multiple human lifetimes.
Join us as we dive deep into the science, challenges, and philosophical questions surrounding these massive space-faring habitats.
What you’ll learn in this video:
**The Structure and Design of a Generation Ship**: Discover how these spacecraft could be built to support life for centuries, from creating artificial gravity to maintaining a self-sustaining ecosystem. We’ll explore the futuristic technologies, such as nuclear fusion engines and advanced life support systems, that could make these ships possible.
**Daily Life Aboard a Generation Ship**: What would day-to-day life look like on a ship traveling through deep space for centuries? We’ll talk about the work, education, and entertainment that would keep future generations engaged. How would people handle the psychological strain of never seeing Earth or their final destination?
**Society and Governance**: How would politics and society evolve in the isolated, confined world of a generation ship? We explore the potential for new social structures, systems of governance, and the ethical dilemmas surrounding generations born aboard who may have never chosen this life.
Whether its pioneering spirit and an urge to explore or a chance to make a fortune or new life out in space, colonization of our solar system may pursue many strategies across many new worlds.
Start Forging New Worlds: https://www.worldanvil.com/isaac-arthur.
Join this channel to get access to perks:
/ @isaacarthursfia.
Visit our Website: http://www.isaacarthur.net.
Join Nebula: https://go.nebula.tv/isaacarthur.
Support us on Patreon: / isaacarthur.
Support us on Subscribestar: https://www.subscribestar.com/isaac-a… Group: / 1,583,992,725,237,264 Reddit:
/ isaacarthur Twitter:
/ isaac_a_arthur on Twitter and RT our future content. SFIA Discord Server:
/ discord Credits: Solar System Colonization Strategies Science & Futurism with Isaac Arthur Episode 316, November 11, 2021 Written, Produced & Narrated by Isaac Arthur Editors: A.T. Long Keith Blockus Sig’unnr Cover Art: Jakub Grygier https://www.artstation.com/jakub_grygier Graphics: Fishy Tree https://www.deviantart.com/fishytree/ Jarred Eagley Jeremy Jozwik https://www.artstation.com/zeuxis_of_… Katie Byrne Ken York
/ ydvisual Kris Holland of www.maficstudios.com Sam McNAmara Sergio Botero https://www.artstation.com/sboterod?f… Space Engine Music Courtesy of Epidemic Sound http://epidemicsound.com/creator.
Facebook Group: / 1583992725237264
Reddit: / isaacarthur.
Twitter: / isaac_a_arthur on Twitter and RT our future content.
SFIA Discord Server: / discord.
Credits:
Solar System Colonization Strategies.
Science & Futurism with Isaac Arthur.
Episode 316, November 11, 2021
Written, Produced & Narrated by Isaac Arthur.
Editors:
A.T. Long.
Keith Blockus.
Sig’unnr.
Cover Art:
Jakub Grygier https://www.artstation.com/jakub_grygier.
Graphics:
Megastructures of the future may range in size from cities to entire galaxies, and must be built to withstand damage and time, but how is this done and what happens when a megastructure dies?
Visit our sponsor, Brilliant: https://brilliant.org/IsaacArthur/
Join this channel to get access to perks:
https://www.youtube.com/channel/UCZFipeZtQM5CKUjx6grh54g/join.
Visit our Website: http://www.isaacarthur.net.
Join Nebula: https://go.nebula.tv/isaacarthur.
Support us on Patreon: https://www.patreon.com/IsaacArthur.
Support us on Subscribestar: https://www.subscribestar.com/isaac-arthur.
Facebook Group: https://www.facebook.com/groups/1583992725237264/
Reddit: https://www.reddit.com/r/IsaacArthur/
Twitter: https://twitter.com/Isaac_A_Arthur on Twitter and RT our future content.
SFIA Discord Server: https://discord.gg/53GAShE
Credits:
Megastructure Death.
Science & Futurism with Isaac Arthur.
Episode 308, September 16, 2021
Written, Produced & Narrated by Isaac Arthur.
Editors:
A.T. Long.
Curt Hartung.
Jason Burbank.
Jerry Guern.
Keith Blockus.
Cover Art:
Jakub Grygier https://www.artstation.com/jakub_grygier.
Graphics:
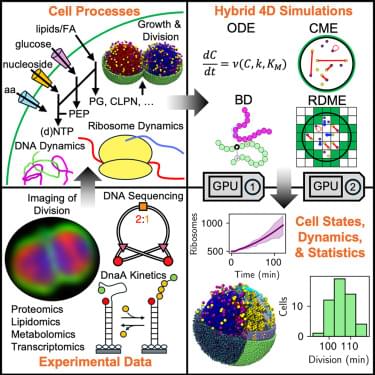
This work represents a fundamental advance in understanding life’s basic principles. By building a cell from the bottom up—specifying every gene, protein, and reaction—researchers can test how life functions with minimal complexity. The simulation serves as a “digital twin” that allows scientists to probe questions impossible to address experimentally, such as how spatial organization affects cellular processes or how subtle parameter changes alter cell cycle timing.
Simulating the complete cell cycle of the minimal cell provides a platform to understand the progression of complete states over time. The spatial heterogeneity of the intracellular environment can strongly affect biochemical reactions that control phenotypes.

Wang P, Zhao D, Lachman HM, Zheng D. Enriched expression of genes associated with autism spectrum disorders in human inhibitory neurons. Transl Psychiatry. 2018;8:13. https://doi.org/10.1038/S41398-017-0058-6