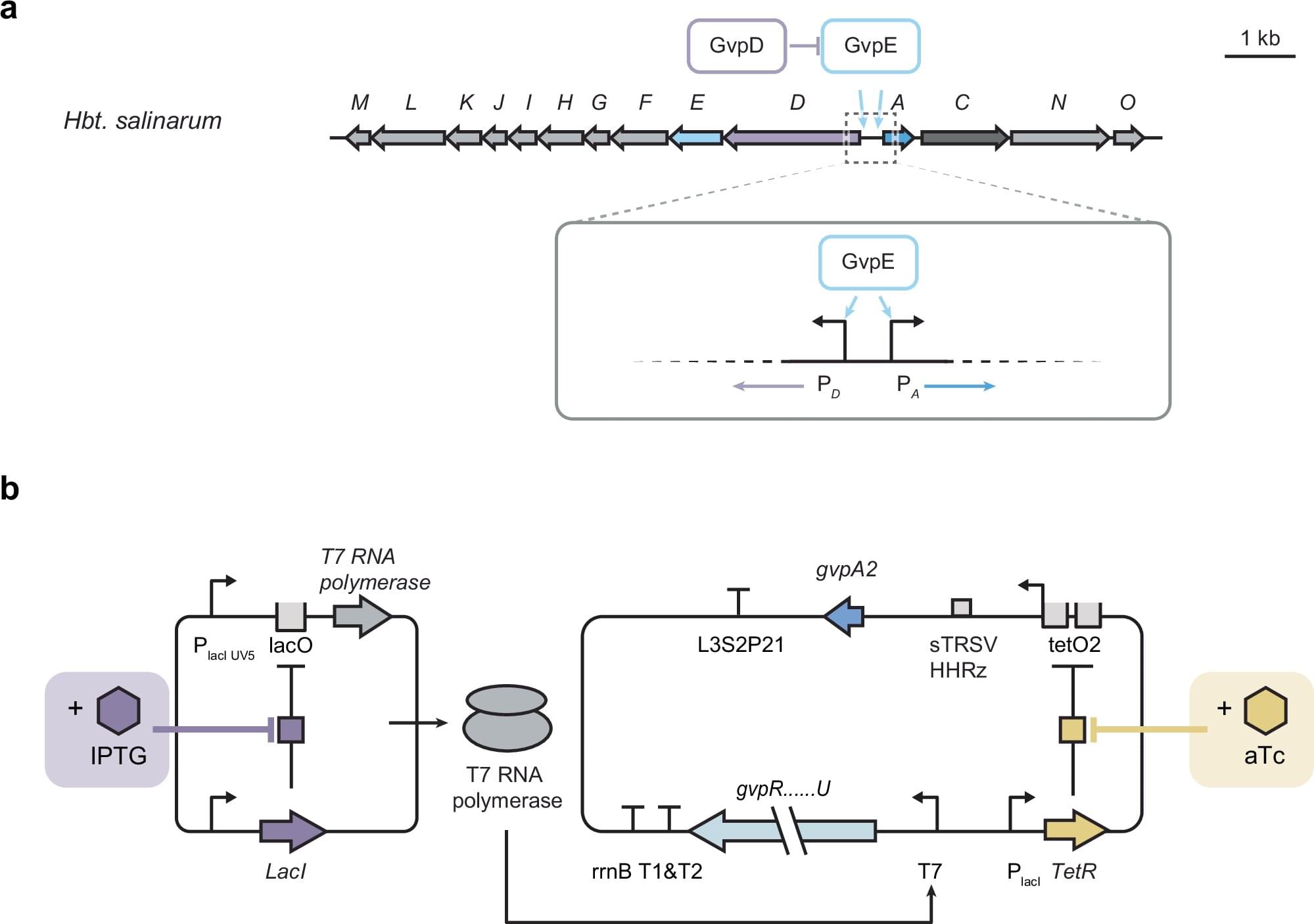
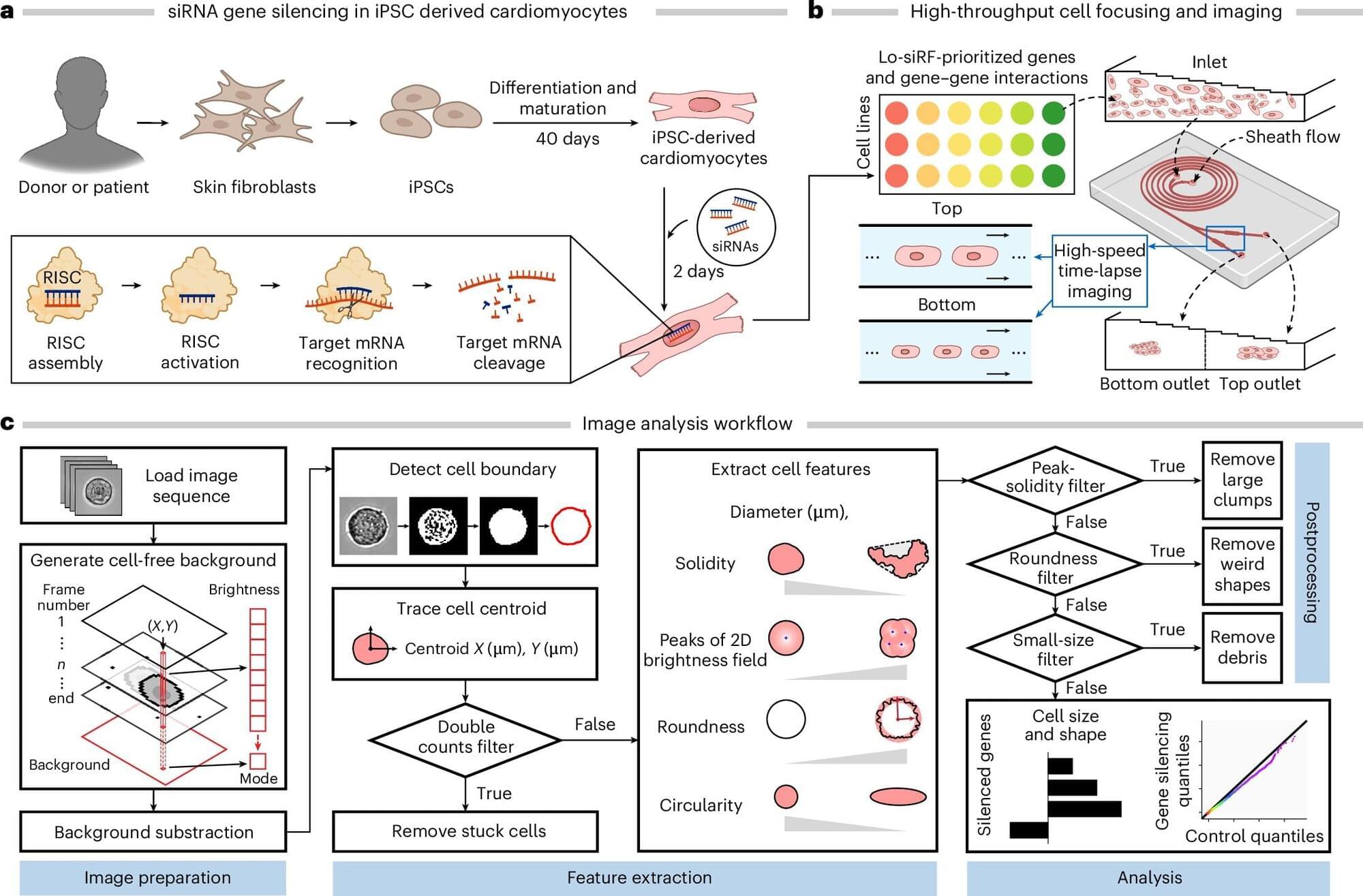
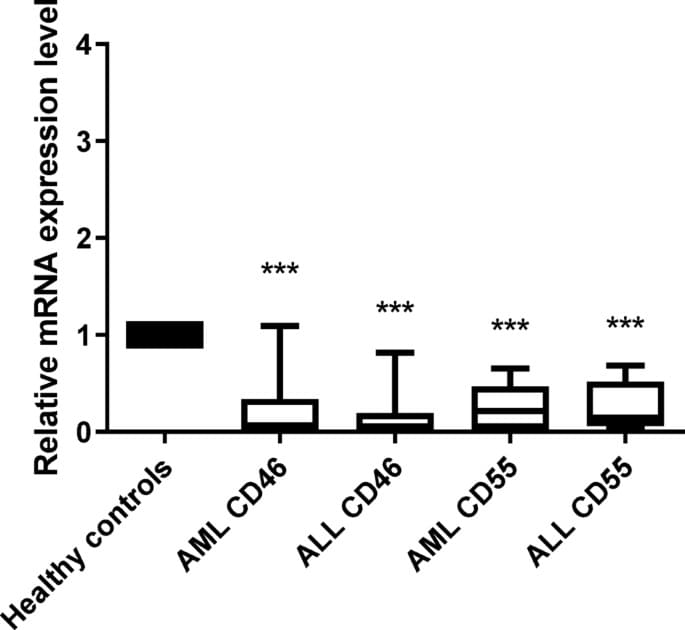
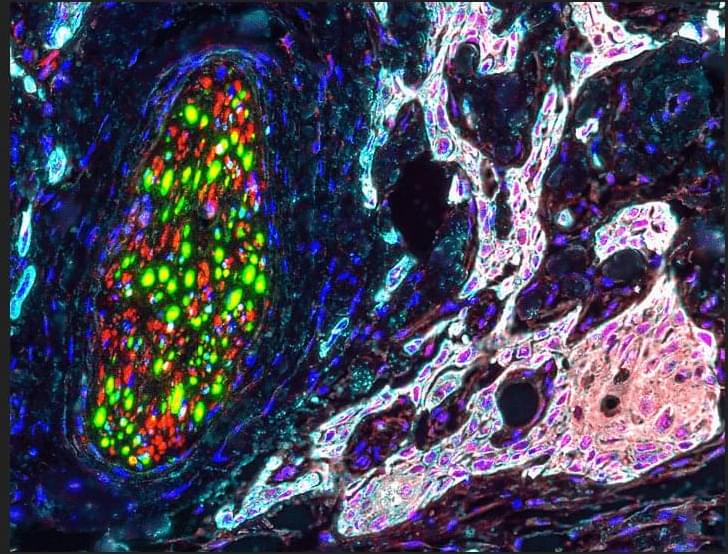
The last few weeks in longevity science have been absolutely unreal. In this episode of Longevity Science News, Emmett Short breaks down 5 bombshell breakthroughs that could reshape the future of human health in 2026 — including an FDA-approved trial aiming to reverse cellular aging, cancer vaccines eliminating brain tumors in days, the regeneration of human teeth, one-shot GLP-1 Ozempic-style gene therapies, and a shocking new discovery linking gut bacteria to multiple sclerosis.
These aren’t sci-fi predictions — these are real developments happening right now in clinical trials, biotech labs, and cutting-edge medical research. If you care about anti-aging, regenerative medicine, epigenetic reprogramming, cancer immunotherapy, GLP-1 weight loss drugs, or the future of human lifespan, this is the episode you don’t want to miss.
Hume Band 20% off with Code LSN20
https://humehealth.com/pages/hume-ban… Huma Band Review: • Best Fitness Tracker For Longevity: Hume B… JOIN LSN Patreon for exclusive access to news, tips and a community of like minded longevity enthusiasts: https://www.patreon.com/user?u=29506604 ✅ Chapters 00:00 – The Longevity Science Explosion 00:48 Hume Band 20% Off 01:02 – Exclusive Interviews 01:43 Bombshell #1: FDA Approves Age Reversal Trial (Yamanaka Factors) 04:40 – Bombshell #2: Cancer’s Worst Month Ever (Vaccines + Immunotherapy) 09:19 – Bombshell #3: The Regeneration Revolution (Cartilage, Teeth, Liver) 11:30 – Bombshell #4: The One-Shot Ozempic Gene Therapy 12:25 – Bombshell #5: Gut Bacteria Linked to Multiple Sclerosis 13:55 – Final Recap + What Breakthrough Comes Next? Links in Script David Sinclair FDA Trial Tweet
https://twitter.com/davidasinclair/status/2
… FDA Greenlights Age Reset Trial (Endpoints) https://endpoints.news/exclusive-fda–… Life Biosciences Epigenetic Reprogramming Video • Reprogramming Human Life — Michael Ringel… mRNA Brain Cancer Vaccine Tweet
just setting up my twttr
— jack (@jack) March 21, 2006
… ANKTIVA Glioblastoma Case Tweet
https://twitter.com/LoriMills4CA42/status/2
… Dr. Patrick Soon-Shiong ANKTIVA Clip Tweet
stalking my coworkers
— crystal (@crystal) March 24, 2006
… Dr. Soon-Shiong Cancer Clip (YouTube) • Patrick Soon-Shiong’s cancer drug Anktiva… MIT/Stanford AbLecs Cancer Breakthrough Tweet
https://twitter.com/ShiningScience/status/2
… Universal mRNA Cancer Vaccine Tweet
https://twitter.com/ShiningScience/status/2
… AI Urine Test for Cancer Detection Tweet
https://twitter.com/Dr_Singularity/status/2
… Akkermansia Gut Bacteria + Immunotherapy Tweet
https://twitter.com/drwilliamli/status/2006
… Cartilage Regeneration Tweet (Liz Parrish)
I feel bad for Pluto. No one likes being demoted.
— David Warner (@hiddeninput) August 24, 2006
… Stanford Cartilage Regeneration Article https://news.stanford.edu/stories/202… Tooth Regrowth Drug Trial Tweet
https://twitter.com/kimmonismus/status/2006
… NewLimit Liver Rejuvenation Tweet
https://twitter.com/byersblake/status/20086
… One-Shot GLP-1 Gene Therapy Thread (Cremieux) https://twitter.com/cremieuxrecueil/status/.… MS Gut Bacteria Breakthrough Video Tweet
trying to get odeo thoughts down
— Ev (@ev) March 23, 2006
… ⚠️ Disclaimer: This video is for educational and informational purposes only and does not constitute medical advice. Consult a qualified clinician before making health or treatment decisions. 🔗 EXCLUSIVE INTERVIEWS & BONUS CONTENT Get extended conversations, deep dives, and behind-the-scenes research ans a YouTube Member Patreon: 👉 / u29506604 YT Membership: 👉
/ @longevitysciencenews PRODUCTION CREDITS ⎺⎺⎺⎺⎺⎺⎺⎺⎺⎺⎺⎺⎺⎺⎺⎺⎺⎺⎺⎺ Executive Producer – Keith Comito @Retromancers Host, Producer, Writer – Emmett Short @emmettshort
Full huma band review: • best fitness tracker for longevity: hume B…
JOIN LSN Patreon for exclusive access to news, tips and a community of like minded longevity enthusiasts: https://www.patreon.com/user?u=29506604
✅ Chapters.