Metabolic plasticity allows cancer cells to adjust their metabolic phenotypes to adapt in hostile environments. There is an urgent need to understand the cross-talk between gene regulation and metabolic pathways underlying cancer metabolic plasticity. We establish a theoretical framework to decode the coupling of gene regulation and metabolic pathways. Our work characterizes a hybrid metabolic state where cells can use both glycolysis and oxidative phosphorylation (OXPHOS) and a possible metabolically inactive state where cells have low activity of both glycolysis and OXPHOS. We show that targeting both OXPHOS and glycolysis may be necessary to eliminate cancer aggressiveness. Our work serves as a platform to target abnormal metabolism in cancer by modulating both genes and metabolic pathways.
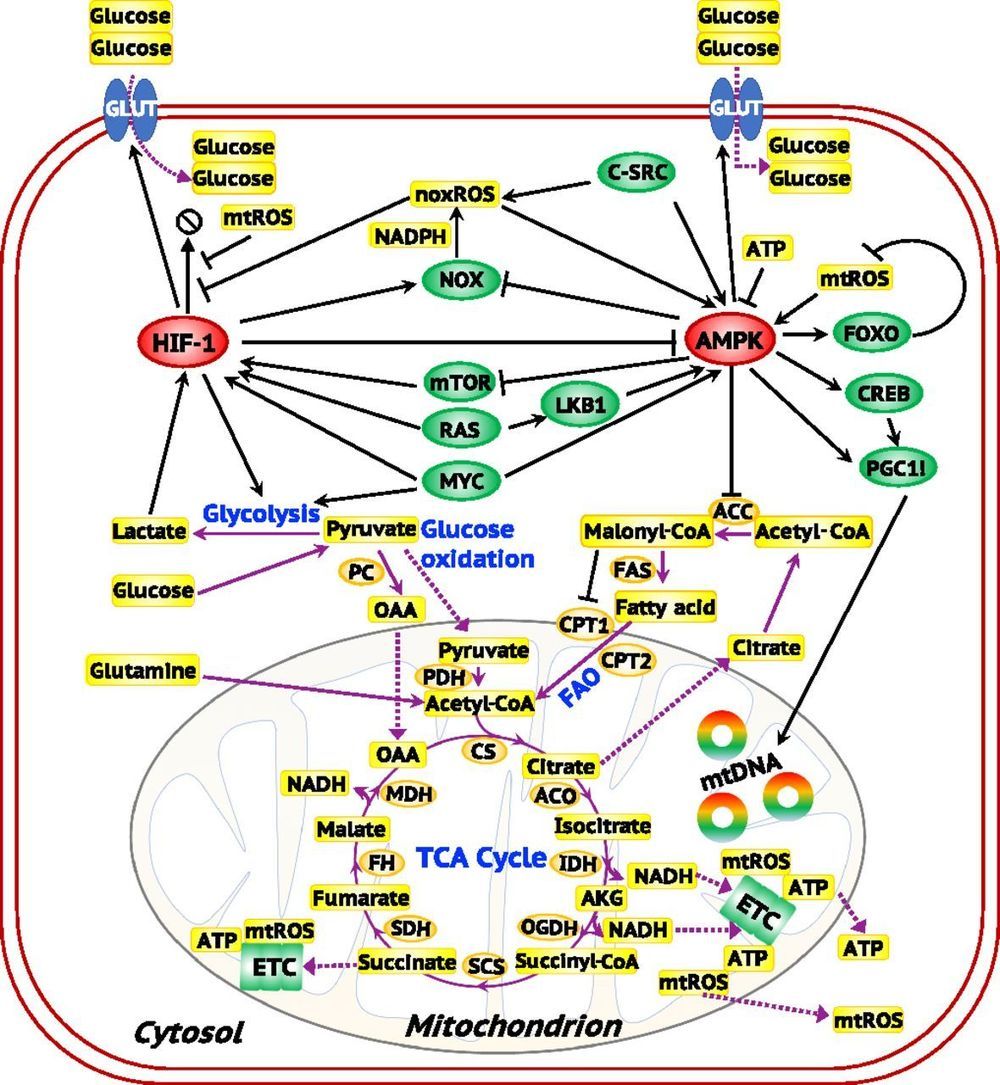
Metabolic plasticity enables cancer cells to switch their metabolism phenotypes between glycolysis and oxidative phosphorylation (OXPHOS) during tumorigenesis and metastasis. However, it is still largely unknown how cancer cells orchestrate gene regulation to balance their glycolysis and OXPHOS activities. Previously, by modeling the gene regulation of cancer metabolism we have reported that cancer cells can acquire a stable hybrid metabolic state in which both glycolysis and OXPHOS can be used. Here, to comprehensively characterize cancer metabolic activity, we establish a theoretical framework by coupling gene regulation with metabolic pathways. Our modeling results demonstrate a direct association between the activities of AMPK and HIF-1, master regulators of OXPHOS and glycolysis, respectively, with the activities of three major metabolic pathways: glucose oxidation, glycolysis, and fatty acid oxidation.