The entirety of the known universe is teeming with an infinite number of molecules. But what fraction of these molecules have potential drug-like traits that can be used to develop life-saving drug treatments? Millions? Billions? Trillions? The answer: novemdecillion, or 1060. This gargantuan number prolongs the drug development process for fast-spreading diseases like COVID-19 because it is far beyond what existing drug design models can compute. To put it into perspective, the Milky Way has about 100 thousand million, or 108, stars.
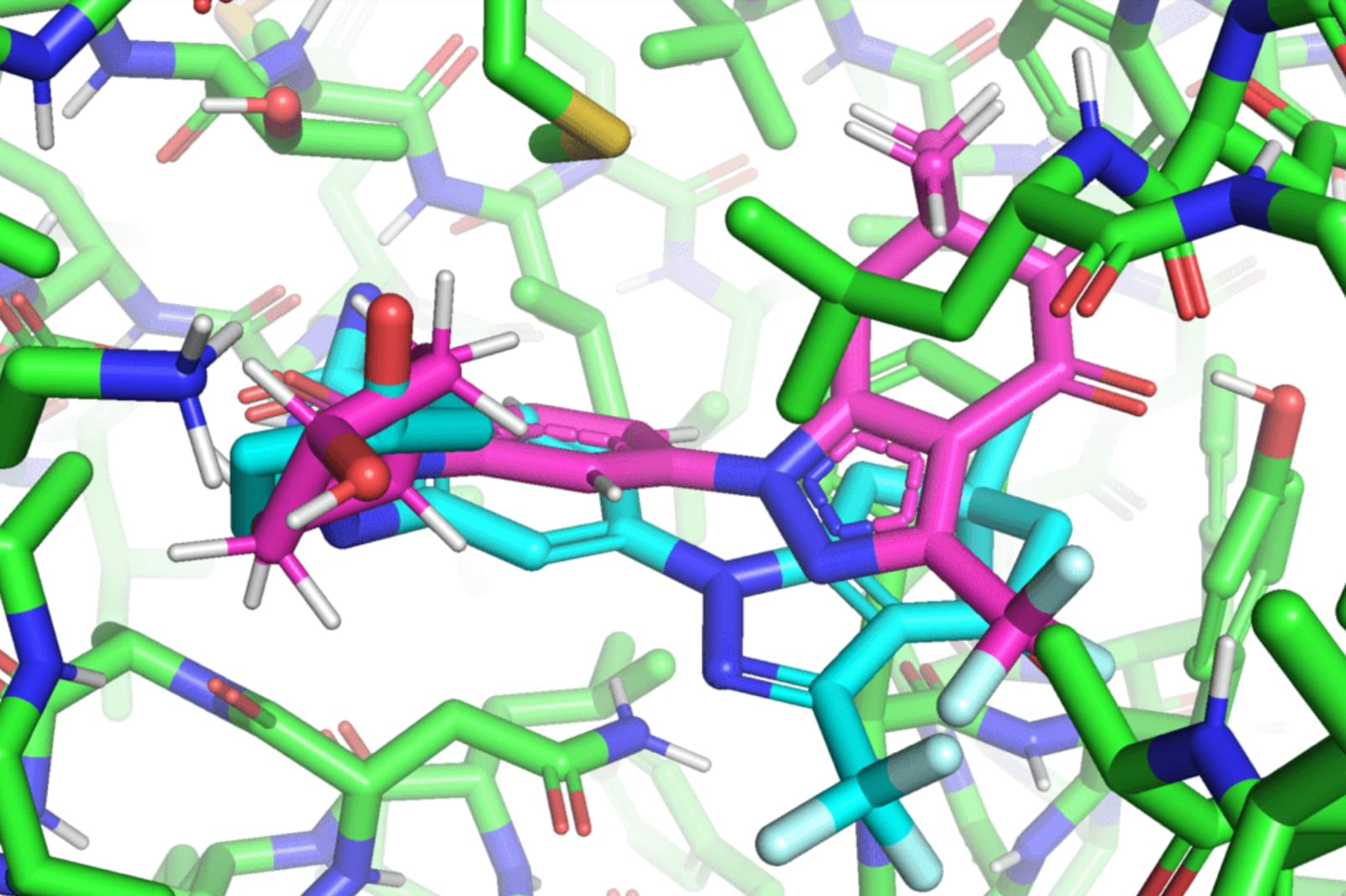
In a paper that will be presented at the International Conference on Machine Learning (ICML), MIT researchers developed a geometric deep-learning model called EquiBind that is 1,200 times faster than one of the fastest existing computational molecular docking models, QuickVina2-W, in successfully binding drug-like molecules to proteins. EquiBind is based on its predecessor, EquiDock, which specializes in binding two proteins using a technique developed by the late Octavian-Eugen Ganea, a recent MIT Computer Science and Artificial Intelligence Laboratory and Abdul Latif Jameel Clinic for Machine Learning in Health (Jameel Clinic) postdoc, who also co-authored the EquiBind paper.
Before drug development can even take place, drug researchers must find promising drug-like molecules that can bind or “dock” properly onto certain protein targets in a process known as drug discovery. After successfully docking to the protein, the binding drug, also known as the ligand, can stop a protein from functioning. If this happens to an essential protein of a bacterium, it can kill the bacterium, conferring protection to the human body.