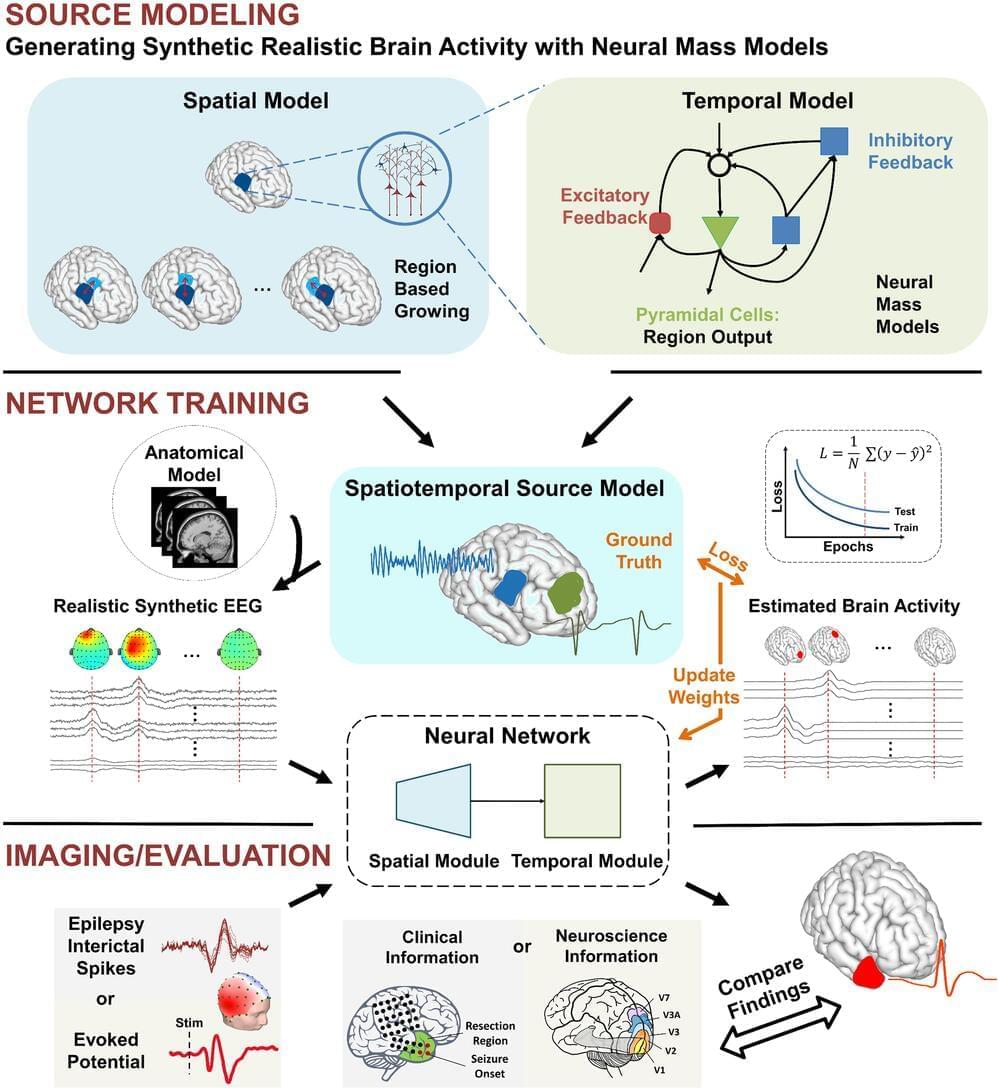
Many efforts have been made to image the spatiotemporal electrical activity of the brain with the purpose of mapping its function and dysfunction as well as aiding the management of brain disorders. Here, we propose a non-conventional deep learning–based source imaging framework (DeepSIF) that provides robust and precise spatiotemporal estimates of underlying brain dynamics from noninvasive high-density electroencephalography (EEG) recordings. DeepSIF employs synthetic training data generated by biophysical models capable of modeling mesoscale brain dynamics. The rich characteristics of underlying brain sources are embedded in the realistic training data and implicitly learned by DeepSIF networks, avoiding complications associated with explicitly formulating and tuning priors in an optimization problem, as often is the case in conventional source imaging approaches. The performance of DeepSIF is evaluated by 1) a series of numerical experiments, 2) imaging sensory and cognitive brain responses in a total of 20 healthy subjects from three public datasets, and 3) rigorously validating DeepSIF’s capability in identifying epileptogenic regions in a cohort of 20 drug-resistant epilepsy patients by comparing DeepSIF results with invasive measurements and surgical resection outcomes. DeepSIF demonstrates robust and excellent performance, producing results that are concordant with common neuroscience knowledge about sensory and cognitive information processing as well as clinical findings about the location and extent of the epileptogenic tissue and outperforming conventional source imaging methods. The DeepSIF method, as a data-driven imaging framework, enables efficient and effective high-resolution functional imaging of spatiotemporal brain dynamics, suggesting its wide applicability and value to neuroscience research and clinical applications.