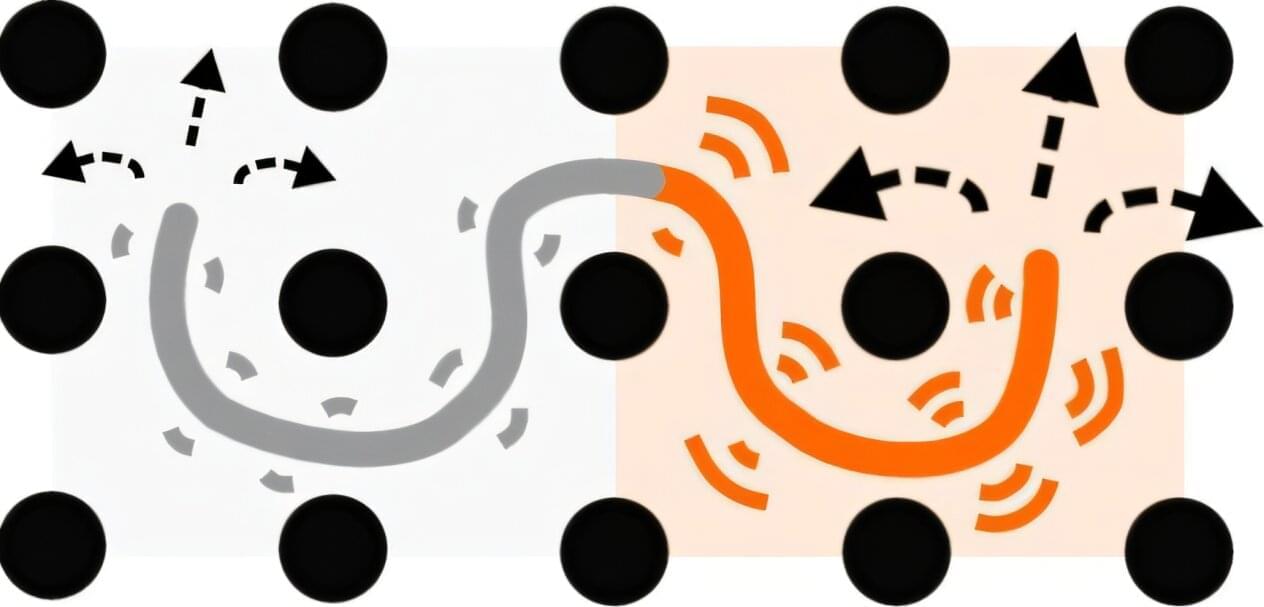
Researchers at the University of Vienna have uncovered a surprising phenomenon: polymer chains with segments that simply fluctuate at different intensities can spontaneously develop directional, persistent motion when densely packed—even though nothing in the system points them in any particular direction. This “entropic tug of war,” driven by fundamental physical constraints, could help explain how DNA organizes and moves inside living cells and may lead to new materials. The study is published in Physical Review X.
“Think of a chain threaded through a dense forest of trees, which represent obstacles posed by the other chains in the system. One end of the chain is being shaken much more vigorously than the other,” explains lead author Jan Smrek from the Faculty of Physics at the University of Vienna. “You might expect it to just wiggle randomly in place. But we found that because the chain has to find its way by going in-between the trees, the difference in shaking intensity creates an imbalance that actually propels the entire chain forward through the forest.”
This analogy can be conferred to a polymer, a large molecule consisting of many units linked together in a long chain, such as DNA. The Viennese research team—Adam Höfler, Iurii Chubak, Christos Likos and Jan Smrek—used computer simulations and analytical theory to show that this directed motion arises purely from topological constraints. When polymer chains are entangled and cannot pass through each other, segments with stronger fluctuations generate larger entropic forces. This creates an imbalance that pushes the entire chain forward along its own contour, with the stronger fluctuating part acting as the “head of the snake” moving through the forest of obstacles.