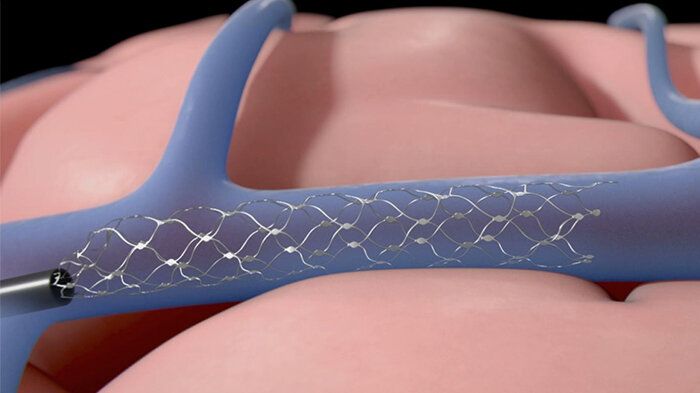
A company that makes an implantable brain-computer interface (BCI) has been given the go-ahead by the Food and Drug Administration to run a clinical trial with human patients. Synchron plans to start an early feasibility study of its Stentrode implant later this year at Mount Sinai Hospital, New York with six subjects. The company said it will assess the device’s “safety and efficacy in patients with severe paralysis.” https://www.engadget.com/fda-brain-computer-interface-clinic…ml?src=rss
A company that makes an implantable has been given the go-ahead by the Food and Drug Administration to run a clinical trial with human patients. Synchron plans to start an early feasibility study of its Stentrode implant later this year at Mount Sinai Hospital, New York with six subjects. The company said it will assess the device’s “safety and efficacy in patients with severe paralysis.”
Synchron received the FDA’s green light ahead of competitors like Elon Musk’s. Before such companies can sell BCIs commercially in the US, they need to prove that the devices work and are safe. The FDA will provide guidance for trials of BCI devices for patients with paralysis or amputation during a webinar on Thursday.
Another clinical trial of Stentrode is underway in Australia. Four patients have received the implant, which is being used “for data transfer from motor cortex to control digital devices,” Synchron said. According to data published in the Journal of NeuroInterventional Surgery, two of the patients were able to control their computer with their thoughts. They completed work-related tasks, sent text messages and emails and did online banking and shopping.